When it comes to sequencing the human genome, "complete" has always been a relative term. The first, deciphered 20 years ago, it included most of the protein-coding regions, but dropped out  approximately 200 million bases of DNA intact, the 8% of the human genome. Even when the additional genomes were "finished," some traits remained out of reach, because repetitive segments of DNA confounded the sequencing technologies of the time. Now, a grassroots international effort has solved those hard-to-read bases, producing the most complete human genome ever.
approximately 200 million bases of DNA intact, the 8% of the human genome. Even when the additional genomes were "finished," some traits remained out of reach, because repetitive segments of DNA confounded the sequencing technologies of the time. Now, a grassroots international effort has solved those hard-to-read bases, producing the most complete human genome ever.
In six articles in Science, the Telomere-to-Telomere Consortium (T2T), named for the end caps of chromosomes, deciphers all but five of hundreds of remaining trouble spots, leaving only 10 million bases and the Y chromosome only roughly known . And today, the T2T consortium announced in a tweet that it has filed a correct assembly sequence of the Y.
“I think we couldn't have imagined that even 5 years ago, certainly not 10 years ago,” says the bioinformatician Ewan Birney, deputy director of the European Molecular Biology Laboratory and part of the original Human Genome Project “It's a tour de force .” The T2T researchers say the newly sequenced traits reveal hot spots for gene evolution and underscore the chaotic history of the human genome. “It really gives us insight into the regions of the genome that have gone unseen,” says Deanna Church, a genomist at Inscripta, a gene-editing company.
Previously indecipherable sequences of the genome that have now become clearly visible include the protective telomeres and dense knobs called centromeres, which typically reside in the middle of each chromosome and help orchestrate its replication. Also almost fully revealed are the short arms of the five chromosomes where the centromeres are angled towards one end. Those short arms were known to contain dozens of genes that code for the backbone of ribosomes, the cell's protein factories.
When Birney, Church and their colleagues presented the first draft of a human genome in 2001, and even after it was "completed" and published in 2004, sequencer machines and genome assembly software could not traverse areas where the DNA sequence contained stretches of bases: the repeats could be skipped too easily or their bases could be linked incorrectly. As sequencing technology has improved and costs have fallen, scientists have reduced the number of gaps or incorrectly assembled sequences, culminating in 2017 with the release of a human genome called GRCh38. With fewer than 1000 gaps, it has become for many the "benchmark" against which other human genomes are compared.
 But Karen Miga and Adam Phillippy wanted to do better. Miga, a geneticist at the University of California, Santa Cruz, wanted to learn the exact sequences of the characteristic "satellite" DNA that helps form centromeres. Meanwhile, Phillippy, a bioinformatician at the National Human Genome Research Institute, was busy harnessing new sequencing technologies that could read very long stretches of DNA, reducing the need to piece together shorter sequences. After meeting at a conference, they joined forces. Then, in 2019, Phillippy reportedly succeeded in sequencing the X chromosome from end to end, inspiring dozens of other researchers to join the cause. “It really took on a life of its own,” Miga says.
But Karen Miga and Adam Phillippy wanted to do better. Miga, a geneticist at the University of California, Santa Cruz, wanted to learn the exact sequences of the characteristic "satellite" DNA that helps form centromeres. Meanwhile, Phillippy, a bioinformatician at the National Human Genome Research Institute, was busy harnessing new sequencing technologies that could read very long stretches of DNA, reducing the need to piece together shorter sequences. After meeting at a conference, they joined forces. Then, in 2019, Phillippy reportedly succeeded in sequencing the X chromosome from end to end, inspiring dozens of other researchers to join the cause. “It really took on a life of its own,” Miga says.
To make the task easier, they decided to use an anonymised cell line derived more than 20 years ago from an unusual growth excised from a woman's uterus: a failed pregnancy called a mole, produced when a sperm entered an egg that lacked its own set of chromosomes. With only the genetic material of sperm, such eggs cannot develop into an embryo, but they can still replicate, especially if the sperm provides an X rather than a Y chromosome. In a boon to the project, both members of the resulting cell line's 23 pairs of chromosomes are identical. This "made a big difference" in eliminating the gaps because sequencers didn't have to sort out the differences between the parental chromosomes, says Robert Waterston, a geneticist at the University of Washington, Seattle, who helped lead the Human Genome Project.
The T2T group combined sequencing technologies, including a so-called nanopore device that could read 100,000 bases at a time and another sequencer that was more accurate but only ran about 10,000 bases at once. A final improvement to the latter method increased accuracy, and together the three approaches were able to eliminate all but five of the final trouble spots. “Just seeing the multiple ways they've gone after this [shows] these are really tough issues,” Waterston says.
The approximately 200 million bases finally in the right order and in the right place include more than 1900 genes, most of them copies of known genes. The researchers cataloged the duplicate regions and mobile elements: genetic material of viruses that have been incorporated into the genome. In sequencing each centromere, they learned that the duplicated regions vary greatly in size, unexpected because these knobs serve the same purpose in each chromosome.
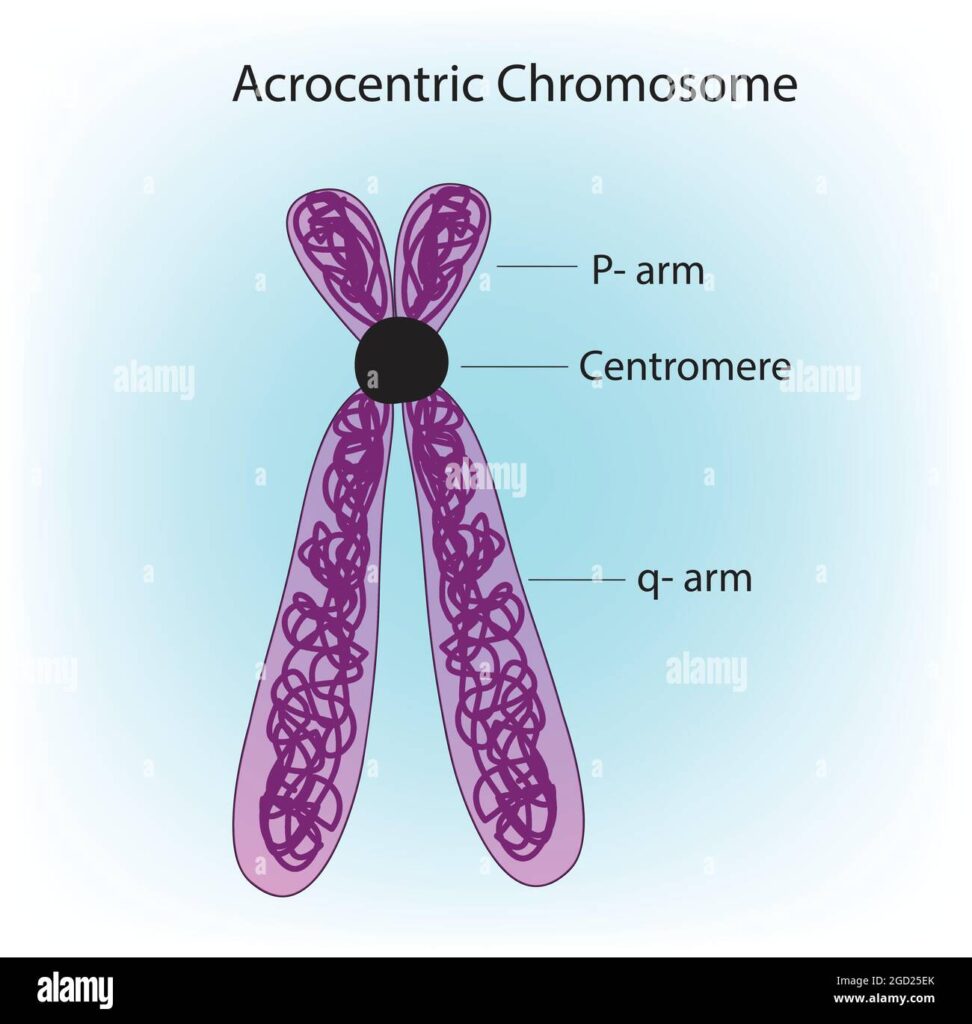 The short arms of the chromosome held another surprise. As expected, they included more copies, 400 in all, of the genes that code for the RNA used to make ribosomes. “This rDNA was the last domino to fall,” as it was the hardest to sequence, Miga says.
The short arms of the chromosome held another surprise. As expected, they included more copies, 400 in all, of the genes that code for the RNA used to make ribosomes. “This rDNA was the last domino to fall,” as it was the hardest to sequence, Miga says.
Short arms are also "just crammed with [more] reps," says Jennifer Gerton, a chromosomal biologist at the Stowers Institute for Medical Research. These include mobile elements, duplicated segments and other types of repetitive DNA, as well as many copies of genes from other parts of the genome. “It's amazing how dynamic the human genome can be,” Church says. At five points along these chromosomes, the resulting jumble is so long that researchers still can't clearly determine the order of the bases, although they have a rough idea of the sequence, Gerton says.
The short arms are likely hotspots for gene evolution, Phillippy notes, as gene copies parked there are free to mutate and take on new functions. The duplication catalog could also shed light on neurological and developmental disorders, which have been linked to variations in the copy number of specific sequences. Chemical changes to DNA in complex repetitive areas are also likely to play a role in the disease, and such changes have been mapped. Because the cell line used lacked a Y chromosome, the T2T team sequenced one from a well-studied genome belonging to Harvard University systems biologist Leonid Peshkin (see sidebar, below).
Despite their latest milestone, human genome sequencers aren't packing their bags. "There's still work to be done," says Human Genome Project co-leader Richard Gibbs, a geneticist at Baylor College of Medicine. He and other researchers point out that the field now needs to obtain similarly complete genomic sequences from a greater diversity of people to look for variations in the short arms and other hard-to-read regions that could play a role in diseases or traits.
The T2T team started by deciphering 70 more genomes, with the goal of 350 from people of different origins. These genomes, sequenced as part of the Human Pangenome Reference Consortium, are more difficult to complete because they do not have identical pairs of chromosomes. So for now, the team has opted for high-quality genomes that place as many bases as possible on their correct chromosomes. Next, the researchers plan to apply all of their methods to Peshkin's entire genome. And, finally, Phillippy says, "We want every genome to be telomere to telomere."
Related articles:
Epigenetic patterns in a complete human genome
Segmental duplications and their variation in a complete human genome




